Project - Mutations in genes causal of white matter disease and dysregulation of lipid metabolism in frontotemporal dementias
Research Project Abstract
Frontotemporal dementia (FTD), a common cause of presenile dementia, is characterized by behavioural and/or language changes and progressive cognitive deficits. Major hurdles in the diagnosis and treatment of the disease are due to the heterogeneity of the underlying brain pathology and differing pathogenic disease mechanisms. Our understanding of FTD has been greatly enhanced by the identification of causal genes that initiate and drive the neurodegenerative process. Mutations in a major FTD gene, Granulin (GRN) are associated with white matter abnormalities via dysregulation of lipid metabolism. White matter plays a key role in the co-ordination, integration and information transfer between various grey matter cortical regions.
Our next-generation sequencing of N = 208 FTD patients has identified deleterious mutations in 20 genes (in 35 patients) that have been reported to cause white matter disease. 13 (65%) of these genes have a direct role in both lipid metabolism and white matter disease.
In this project, we will confirm the causal role of the pathogenic mutations in these genes in cellular models and in vivo, with a focus on the impact of these genes on the lipid metabolism from central and peripheral tissue, and their corresponding changes in white matter. In order to improve diagnostic accuracy of this heterogeneous disorder, we will compare our findings to other disease groups including sporadic FTD cases and carriers of other FTD gene mutations such as in MAPT and C9orf72.
Project funded from 2019-202
Challenges within the field
Current clinical, imaging and biomarker needs - The identification of accurate clinical phenotypes is required for optimal treatment strategies. While this has been achieved for AD, comprehensive data are now required for the non-AD dementias, such as FTD. The identification of the full spectrum of FTD causative genes, and the elucidation of their pathogenic mechanisms and their relation to neuroimaging and peripheral biomarkers, will ensure significant advances in the diagnosis of FTD.
Research Project Description
Aim A: Determine the functional basis of FTD - white matter disease genes and impact of mutations in cell culture models – We will confirm whether white matter disease mutations have significant impact on their respective biological pathways, as well as on key FTD neuropathological molecules such as tau and TDP-43. We will use an established oligodendrocyte cell line and induced pluripotent stem cells (iPSCs) that have been differentiated into multiple cellular phenotypes.
Aim B: Perform detailed lipidomic analyses on brain tissue of mutation carriers and sporadic cases - Lipidomic profiles will be generated by mass spectrometry from both grey and white matter of brain regions that typically differ in the severity of FTD pathology in mutation carriers and sporadic cases. Multivariate analyses will examine effects of age, gender, disease severity and pathology.
Aim C: Determine the magnetic resonance imaging correlates in FTD - white matter disease mutation carriers and their overlap with sporadic FTD cases and neurological controls - Utilizing a cohort of mutation carriers with longitudinal data, we will examine the effects of these mutations on changes in white matter structural integrity and functional connectivity. We will determine the best imaging technology to identify and quantify the alterations in white matter in these patients, and to determine whether these changes are also associated with sporadic FTD.
Aim D: Identification of lipid biomarker panel that is accessible from peripheral tissue and will track disease state and severity - Utilizing the same cohort of mutation carriers with longitudinal serum samples, we will perform a thorough screen of lipids to identify all major classes of lipids. We will employ standard statistical approaches and unsupervised machine learning algorithms to identify lipids and white matter changes capable of discriminating both mutation carriers and sporadic cases, and which can be used to improve diagnostic accuracy.
Research Objectives
- Discovery and functional characterisation of genetic variants associated with multiple neurodegenerative diseases.
- Multi-disciplinary projects involving healthy ageing, neurpyschiatric diseases and cancer.
- Develop in vitro protocols that will allow rapid functional screening of genetic variants.
- Translate genetic findings to clinically useful neuroimaging and blood-based biomarkers (based on nucleic acids (cell free DNAs, RNA molecules and lipids).
- Establish next generation sequencing and bioinformatics platform to enhance and translate genetic research.
We wish to become world leaders in the field of neurodegenetics and epigenetics, leveraging the expertise of the world class team of research scientists and clinicians at ForeFront.
Key Publications from this project
- Oyston L..Kwok JB, Dobson-Stone C. Reply: CYLD variants in frontotemporal dementia associated with severe memory impairment in a Portuguese cohort. Brain (accepted for publication 28/04/2020).
- Shin J.. Kwok JB.. neuroCHARGE Working Group. Global and Regional Development of the Human Cerebral Cortex: Molecular Architecture and Occupational Aptitudes. Cereb Cortex. 2020 Mar 20. pii: bhaa035. doi: 10.1093/cercor/bhaa035. [Epub ahead of print].
- Grasby KL..Kwok JB.. Medland SE; Enhancing NeuroImaging Genetics through Meta-Analysis Consortium (ENIGMA)—Genetics working group. The genetic architecture of the human cerebral cortex. Science. 2020 Mar 20;367(6484). pii: eaay6690. doi: 10.1126/science.aay6690.
- Dobson-Stone C.. Kwok JB. CYLD is a causative gene for frontotemporal dementia - amyotrophic lateral sclerosis. Brain 2020 143(3):783-799. doi: 10.1093/brain/awaa039.
- Vallerga CL.. Kwok JB.. Gratten J. Analysis of DNA methylation associates the cystine-glutamate antiporter SLC7A11 with risk of Parkinson's disease. Nat Commun. 2020 Mar 6;11(1):1238. doi: 10.1038/s41467-020-15065-7.
- JiaT..Kwok KB.. Desrivières S. Epigenome-wide meta-analysis of blood DNA methylation and its association with subcortical volumes: findings from the ENIGMA Epigenetics Working Group. Mol Psychiatry. 2019 Dec 6. doi: 10.1038/s41380-019-0605-z. [Epub ahead of print].
- McCann EP.. Kwok JBJ.. Yang S. Genetic and immunopathological analysis of CHCHD10 in Australian amyotrophic lateral sclerosis and frontotemporal dementia and transgenic TDP-43 mice. J Neurol Neurosurg Psychiatry. 2020 Feb;91(2):162-171.
- Satizabal CL.. Kwok..JB.. Ikram MA. Genetic architecture of subcortical brain structures in 38,851 individuals. Nat Genet. 2019 Nov;51(11):1624-1636. doi: 10.1038/s41588-019-0511-y.
- Zhang Q..Kwok JB..Visscher PM. Improved precision of epigenetic clock estimates across tissues and its implication for biological ageing. Genome Med. 2019 Aug 23;11(1):54. doi: 10.1186/s13073-019-0667-1.
- Revelas M.. Kwok.. Mather KA. Exceptional Longevity and Polygenic Risk for Cardiovascular Health. Genes (Basel). 2019 Mar 18;10(3). pii: E227. doi: 10.3390/genes10030227.
- Chauhan G .. Kwok JB.. Neurology Working Group of the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium. Genetic and lifestyle risk factors for MRI-defined brain infarcts in a population-based setting. Neurology. 2019 Jan 16. pii: 10.1212/WNL.0000000000006851. doi: 10.1212/WNL.0000000000006851.
Key Findings
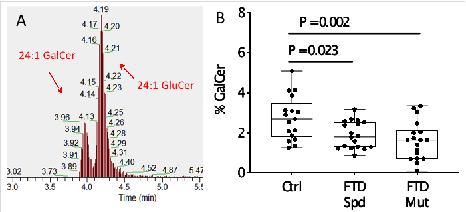
Identification of a blood-based biomarker for myelin integrity in the brain. The tissue of origin of gold-standard biomarkers for neurodegenerative diseases, such as beta-amyloid peptide, cannot be determined using current technologies. In contrast, this can be done for the hexosylceramide class of lipids using a modified mass spectrometry assay to differentiate between brain-derived stereoisomer galactosyl-(GalCer) ceramides from liver-derived glucosyl-(GluCer) species. In collaboration with A/Prof A Don (Centenary Institute), we have demonstrated that plasma C24:1 galactosylceramide was downregulated in WM disease gene mutation carriers compared with neurological controls or sporadic FTD cases. My research will now determine the consequences of these mutations and their association with multi-modal data such as neuropsychiatric phenotypes, disease onset and severity, WM tractography or oligodendrocyte numbers in brain tissue. This will provide robust evidence of the biological and clinical significance of these lipid species and drive the development of a cheap and non-invasive biomarker.
